Using Rbind for Reading Files With Id
Welcome! If you want to beginning diving into data science and statistics, then information frames, CSV files, and R volition be essential tools for you. Let's see how you can employ their amazing capabilities.
In this commodity, you volition learn:
- What CSV files are and what they are used for.
- How to create CSV files using Google Sheets.
- How to read CSV files in R.
- What Data Frames are and what they are used for.
- How to access the elements of a data frame.
- How to alter a data frame.
- How to add together and delete rows and columns.
Nosotros will apply RStudio, an open-source IDE (Integrated Evolution Surroundings) to run the examples.
Let's brainstorm! ✨
🔹 Introduction to CSV Files
CSV (Comma-separated Values) files can be considered one of the building blocks of data assay because they are used to shop data represented in the grade of a table.
In this file, values are separated by commas to represent the dissimilar columns of the table, similar in this instance:
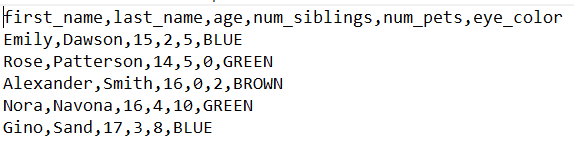
We volition generate this file using Google Sheets.
🔸 How to Create a CSV File Using Google Sheets
Let'southward create your offset CSV file using Google Sheets.
Pace 1: Go to the Google Sheets Website and click on "Go to Google Sheets":
💡 Tip: Yous can access Google Sheets past clicking on the button located at the superlative-correct edge of Google's Home Page:
If we zoom in, nosotros see the "Sheets" button:
💡 Tip: To use Google Sheets, you need to have a Gmail account. Alternatively, you tin can create a CSV file using MS Excel or another spreadsheet editor.
Y'all will come across this panel:
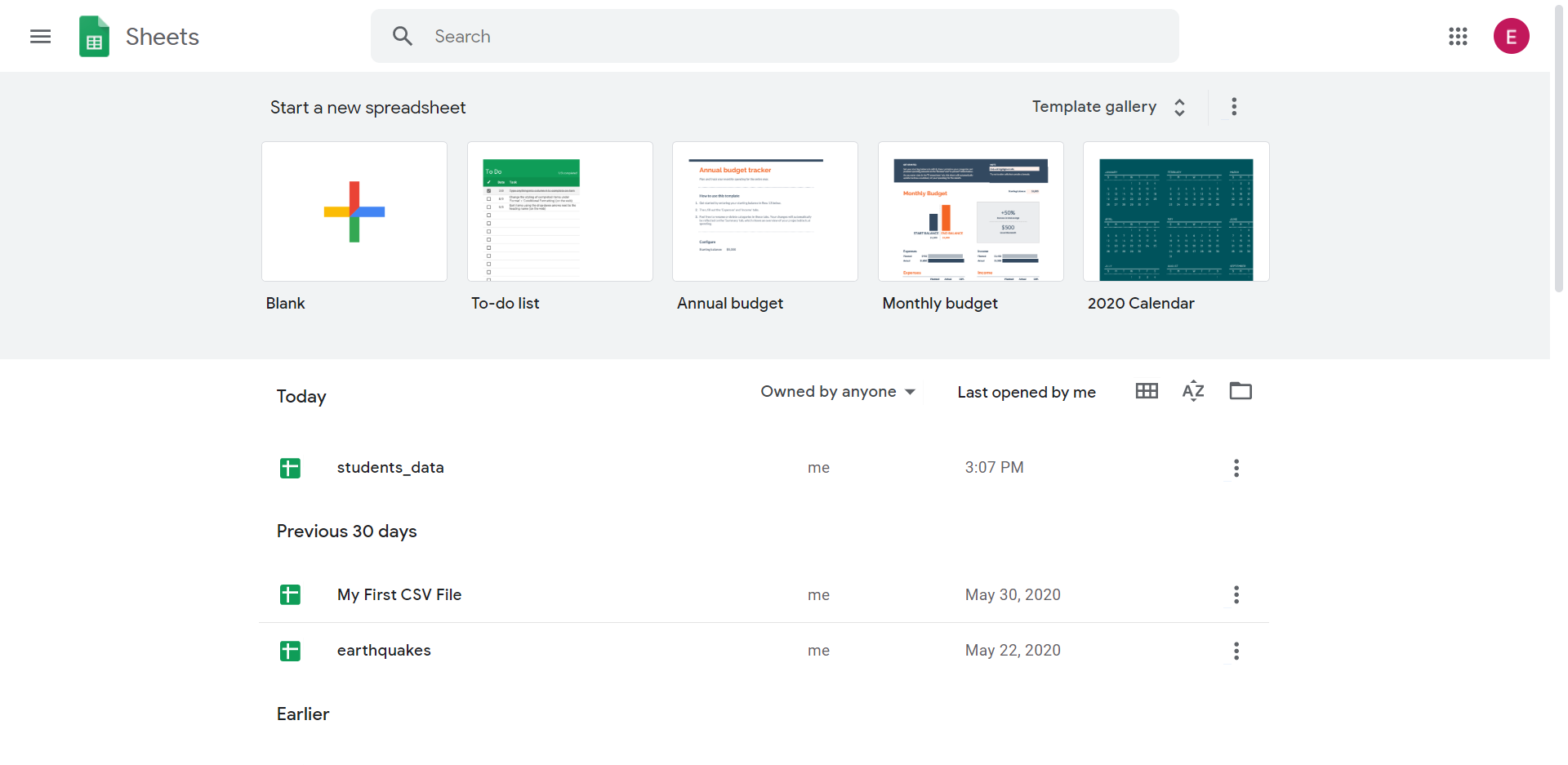
Step 2: Create a blank spreadsheet by clicking on the "+" push button.
At present you take a new empty spreadsheet:
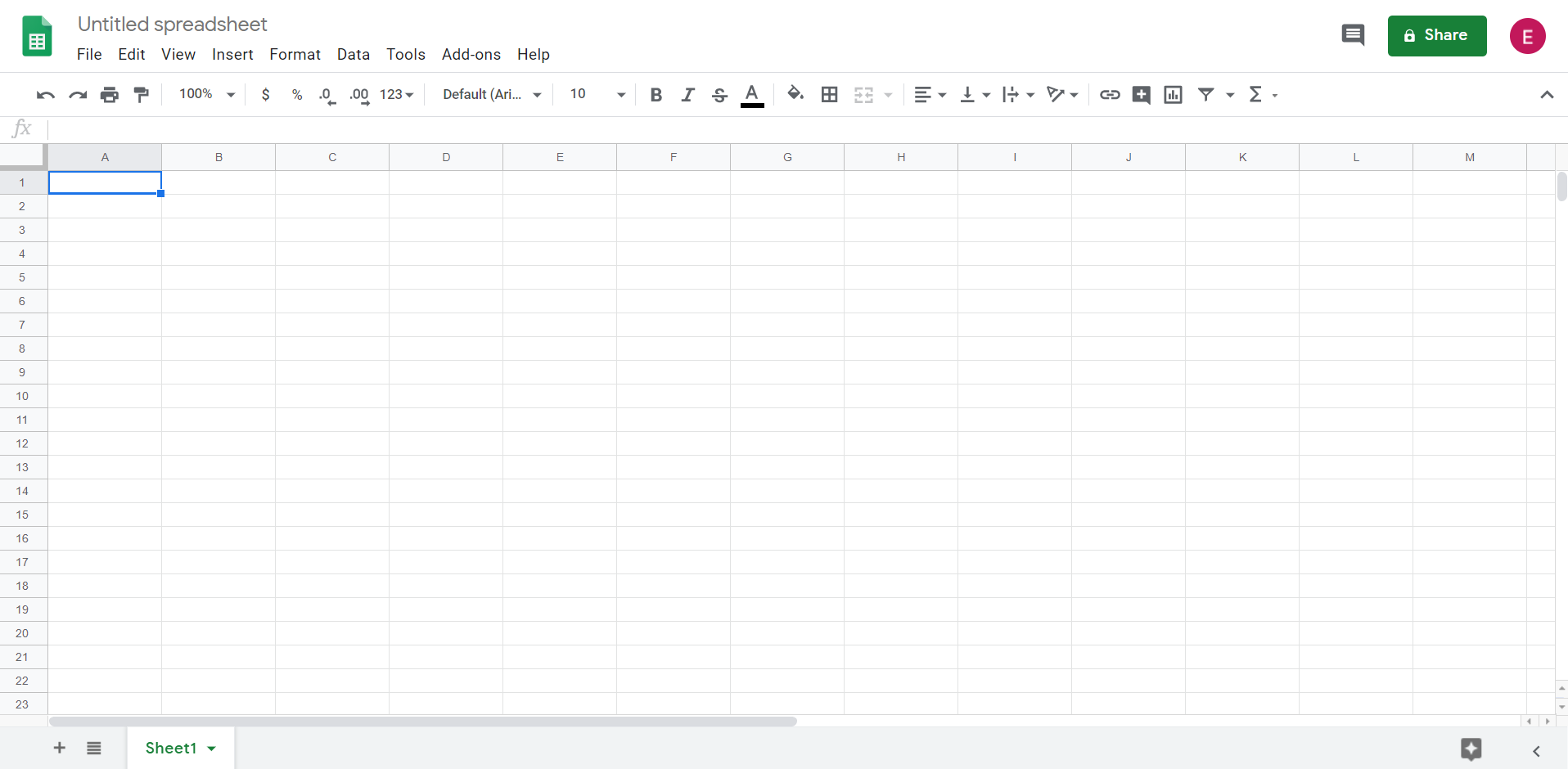
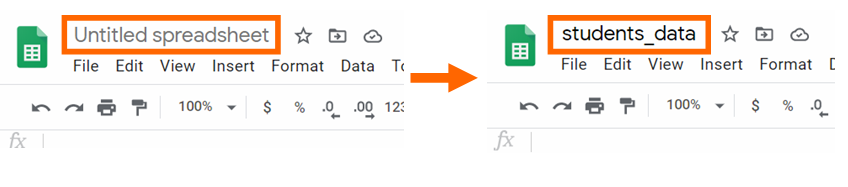
Pace 3: Change the name of the spreadsheet to students_data. We volition need to use the name of the file to work with information frames. Write the new name and click enter to confirm the change.
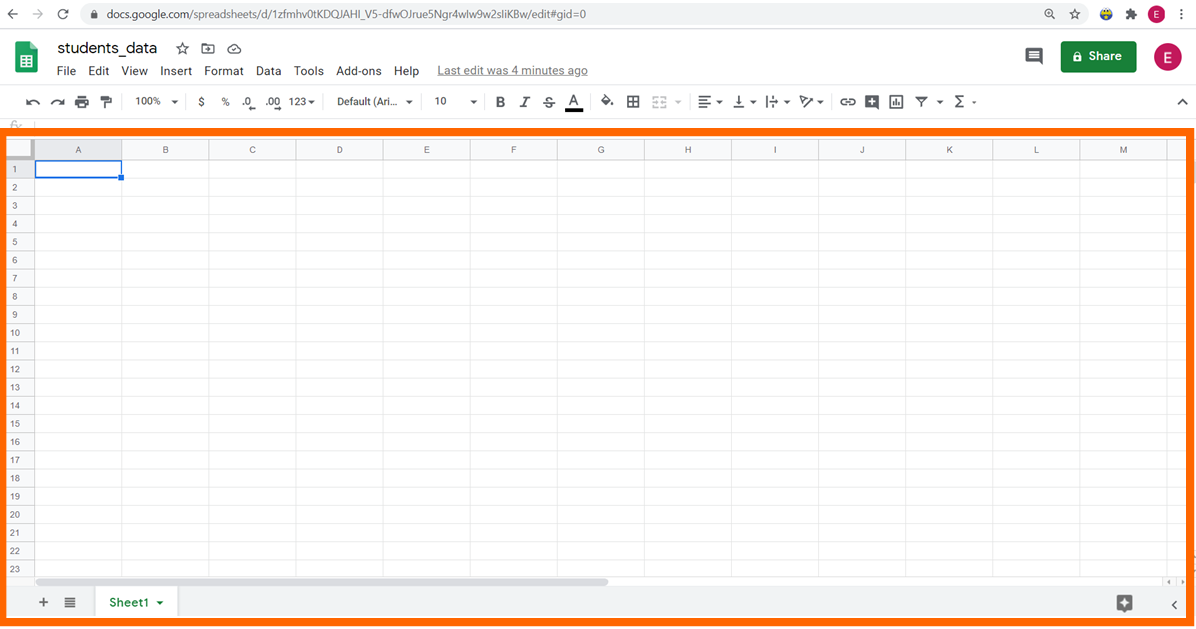
Footstep four: In the first row of the spreadsheet, write the titles of the columns.
When you import a CSV file in R, the titles of the columns are called variables. We volition ascertain 6 variables: first_name, last_name, age, num_siblings, num_pets, and eye_color, equally you can see right here below:
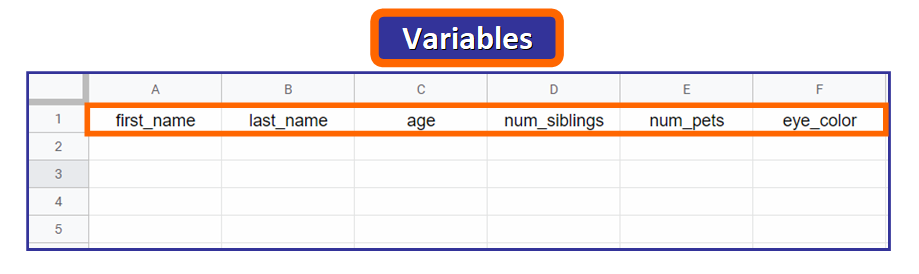
💡 Tip: Observe that the names are written in lowercase and words are separated with an underscore. This is non mandatory, only since you volition demand to access these names in R, information technology'southward very common to utilize this format.
Stride 5: Enter the data for each ane of the columns.
When you read the file in R, each row is chosen an observation, and information technology corresponds to data taken from an individual, animal, object, or entity that we collected data from.
In this instance, each row corresponds to the information of a student:
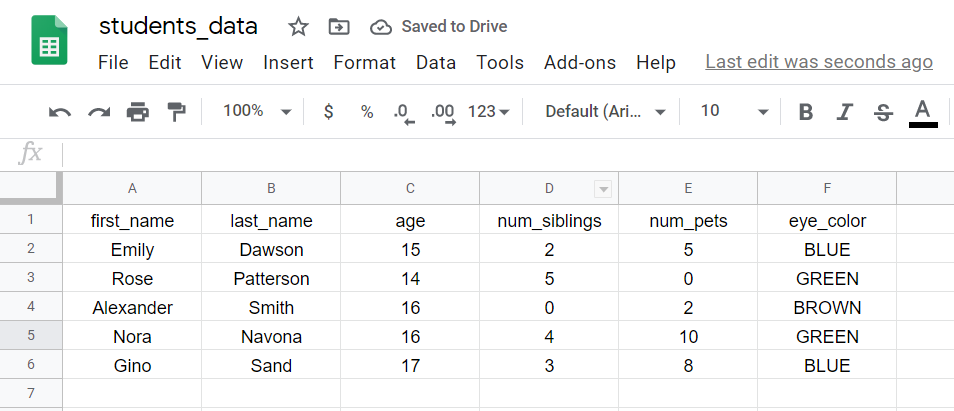
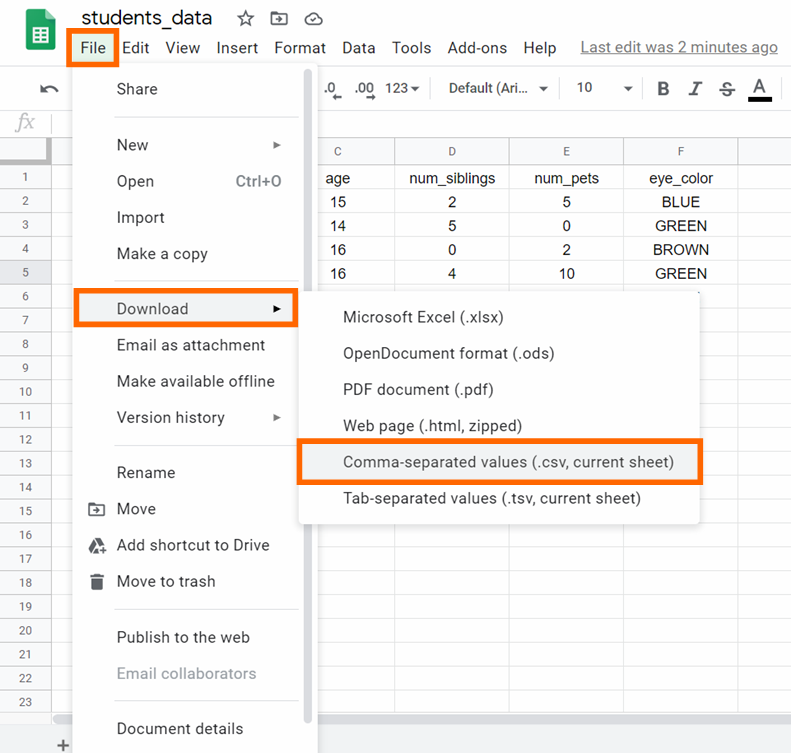
Step six: Download the CSV file past clicking on File -> Download -> Comma-separated values, as you lot tin run into beneath:
Step 7: Rename the file CSV file. You will need to remove "Sheet1" from the default proper noun because Google Canvass volition automatically add this to the name of the file.

Great work! At present yous have your CSV file and information technology's time to start working with it in R.
🔹 How to Read a CSV file in R
In RStudio, the first step before reading a CSV file is making sure that your current working directory is the directory where the CSV file is located.
💡 Tip: If this is not the case, you lot volition demand to use the full path to the file.
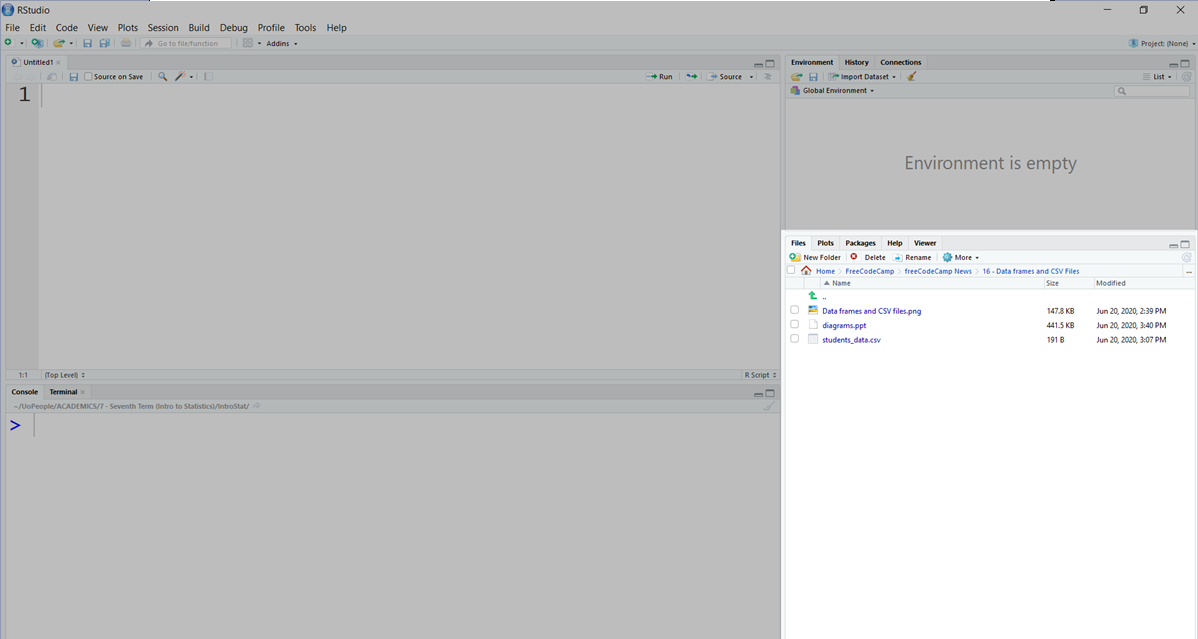
Change Current Working Directory
You can change your current working directory in this panel:
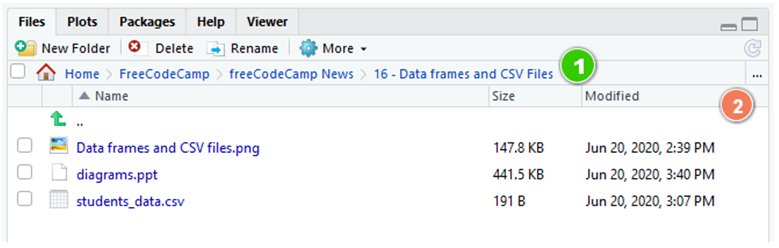
If we zoom in, you tin see the current path (1) and select the new i by clicking on the ellipsis (...) button to the right (2):
💡 Tip: Yous can also check your current working directory with getwd() in the interactive panel.
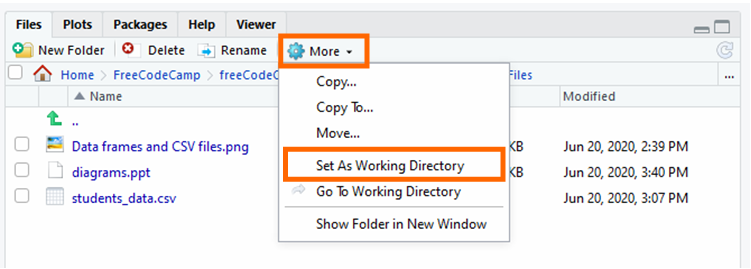
So, click "More than" and "Set up Equally Working Directory".
Read the CSV File
One time you accept your current working directory set up, you tin read the CSV file with this command:

In R code, we accept this:
> students_data <- read.csv("students_data.csv") 💡 Tip: Nosotros assign it to the variable students_data to access the data of the CSV file with this variable. In R, we can divide words using dots ., underscores _, UpperCamelCase, or lowerCamelCase.
Later running this command, you lot will run into this in the top right panel:
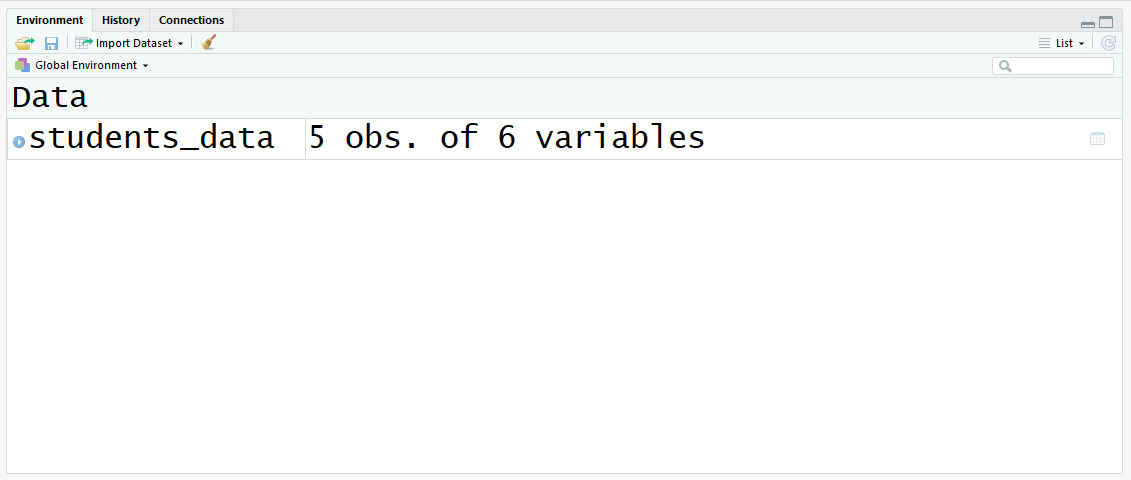
Now you have a variable defined in the environs! Let'south encounter what data frames are and how they are closely related to CSV files.
🔸 Introduction to Data Frames
Data frames are the standard digital format used to store statistical data in the form of a table. When you lot read a CSV file in R, a data frame is generated.
We can ostend this by checking the type of the variable with the class part:
> class(students_data) [1] "data.frame" It makes sense, right? CSV files contain data represented in the form of a table and data frames represent that tabular data in your code, so they are deeply connected.
If you lot enter this variable in the interactive console, y'all will see the content of the CSV file:
> students_data first_name last_name age num_siblings num_pets eye_color 1 Emily Dawson fifteen ii 5 BLUE 2 Rose Patterson 14 5 0 GREEN three Alexander Smith 16 0 ii BROWN four Nora Navona 16 4 10 GREEN 5 Gino Sand 17 3 8 Blue More than Information About the Data Frame
You have several different alternatives to come across the number of variables and observations of the data frame:
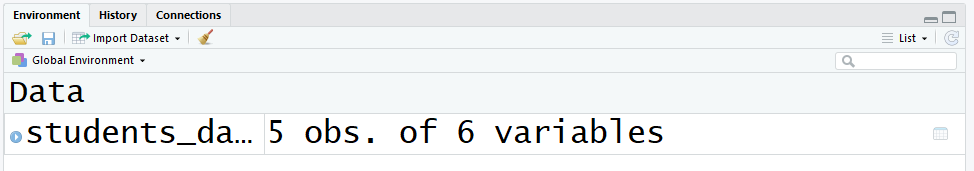
- Your first selection is to expect at the top right console that shows the variables that are currently defined in the environment. This information frame has 5 observations (rows) and 6 variables (columns):
- Another alternative is to utilize the functions
nrowandncolin the interactive panel or in your plan, passing the data frame as argument. We get the same results: five rows and 6 columns.
> nrow(students_data) [ane] 5 > ncol(students_data) [1] half-dozen - You tin can likewise see more data about the information frame using the
strfunction:
> str(students_data) 'data.frame': five obs. of 6 variables: $ first_name : Factor w/ five levels "Alexander","Emily",..: two 5 1 four 3 $ last_name : Factor w/ 5 levels "Dawson","Navona",..: i iii 5 ii 4 $ historic period : int 15 xiv 16 xvi 17 $ num_siblings: int ii v 0 4 iii $ num_pets : int five 0 2 10 8 $ eye_color : Factor w/ three levels "BLUE","Brownish",..: ane iii 2 3 ane This function (applied to a information frame) tells you:
- The number of observations (rows).
- The number of variables (columns).
- The names of the variables.
- The data types of the variables.
- More information about the variables.
You tin come across that this function is really great when y'all want to know more than near the data that yous are working with.
💡 Tip: In R, a "Factor" is a qualitative variable, which is a variable whose values represent categories. For example, eye_color has the values "Blue", "BROWN", "Greenish" which are categories, so as you can see in the output of str to a higher place, this variable is automatically defined as a "factor" when the CSV file is read in R.
🔹 Information Frames: Key Operations and Functions
At present you know how to see more information about the data frame. But the magic of data frames lies in the amazing capabilities and functionality that they offer, and so permit'south see this in more than detail.
How to Access A Value of a Data Frame
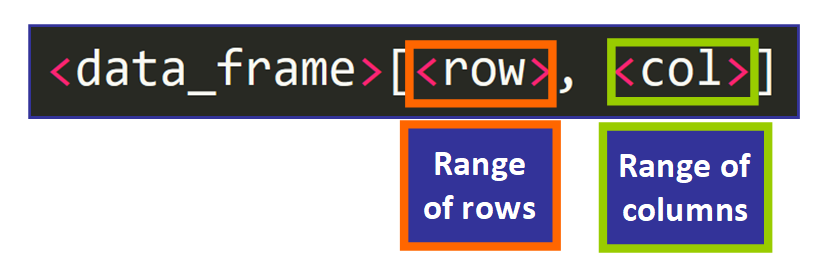
Data frames are similar matrices, and then you tin admission private values using two indices surrounded by square brackets and separated by a comma to point which rows and which columns you would similar to include in the issue, similar this:

For example, if we want to admission the value of eye_color (column 6) of the fourth student in the data (row 4):
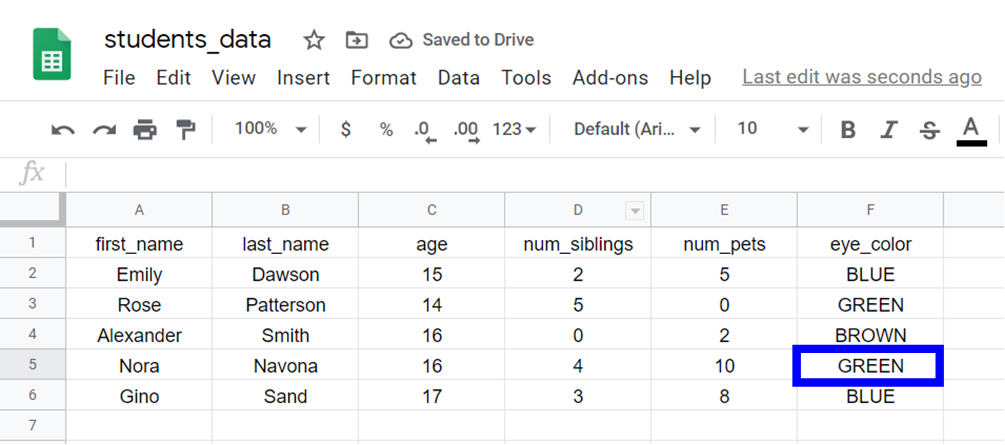
We need to use this control:
> students_data[4, 6] 💡 Tip: In R, indices starting time at 1 and the get-go row with the names of the variables is not counted.
This is the output:
[1] Light-green Levels: BLUE BROWN GREEN You tin can run across that the value is "Green". Variables of type "factor" accept "levels" that stand for the unlike categories or values that they can take. This output tells united states of america the levels of the variable eye_color.
How to Access Rows and Columns of a Data Frame
Nosotros can also use this syntax to admission a range of rows and columns to become a portion of the original matrix, like this:
For example, if we want to get the historic period and number of siblings of the third, fourth, and fifth student in the list, we would use:
> students_data[3:five, iii:four] age num_siblings 3 16 0 iv 16 4 5 17 3 💡 Tip: The bones syntax to ascertain an interval in R is <start>:<end>. Notation that these indices are inclusive, then the third and fifth elements are included in the example in a higher place when we write 3:five.
If nosotros want to get all the rows or columns, nosotros simply omit the interval and include the comma, similar this:
> students_data[3:v,] first_name last_name age num_siblings num_pets eye_color 3 Alexander Smith sixteen 0 ii Brown 4 Nora Navona sixteen 4 10 GREEN five Gino Sand 17 3 8 Bluish Nosotros did not include an interval for the columns afterward the comma in students_data[3:v,], and so we get all the columns of the information frame for the three rows that we specified.
Similarly, nosotros can go all the rows for a specific range of columns if nosotros omit the rows:
> students_data[, one:iii] first_name last_name age one Emily Dawson 15 2 Rose Patterson 14 three Alexander Smith xvi 4 Nora Navona 16 5 Gino Sand 17 💡 Tip: Detect that you yet need to include the comma in both cases.
How to Access a Column
At that place are three ways to access an entire cavalcade:
- Option #1: to access a column and return it every bit a data frame, you can employ this syntax:

For case:
> students_data["first_name"] first_name one Emily 2 Rose 3 Alexander 4 Nora 5 Gino - Option #2: to get a column as a vector (sequence), you can apply this syntax:

💡 Tip: Observe the employ of the $ symbol.
For case:
> students_data$first_name [one] Emily Rose Alexander Nora Gino Levels: Alexander Emily Gino Nora Rose - Option #3: You can as well apply this syntax to get the cavalcade as a vector (see beneath). This is equivalent to the previous syntax:
> students_data[["first_name"]] [1] Emily Rose Alexander Nora Gino Levels: Alexander Emily Gino Nora Rose How to Filter Rows of a Data Frame
You can filter the rows of a data frame to get a portion of the matrix that meets certain weather.
For this, we use this syntax, passing the condition as the starting time element inside square brackets, so a comma, and finally leaving the second element empty.

For case, to get all rows for which students_data$historic period > sixteen, we would apply:
> students_data[students_data$historic period > 16,] first_name last_name age num_siblings num_pets eye_color 5 Gino Sand 17 iii 8 BLUE Nosotros go a information frame with the rows that encounter this status.
Filter Rows and Choose Columns
You can combine this condition with a range of columns:
> students_data[students_data$age > xvi, three:six] age num_siblings num_pets eye_color 5 17 three viii Bluish Nosotros get the rows that encounter the status and the columns in the range 3:vi.
🔸 How to Modify Information Frames
You tin can modify individual values of a information frame, add together columns, add rows, and remove them. Permit's run across how you can practise this!
How to Change A Value
To modify an individual value of the data frame, yous need to use this syntax:
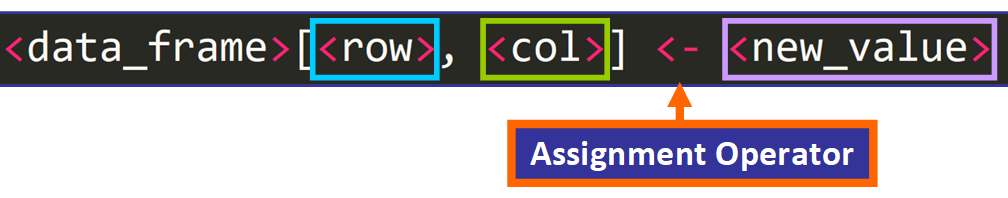
For case, if we want to change the value that is currently at row four and column 6, denoted in bluish correct here:

We need to use this line of code:
students_data[4, 6] <- "Brown" 💡 Tip: You can also use = as the assignment operator.
This is the output. The value was inverse successfully.

💡 Tip: Retrieve that the first row of the CSV file is not counted as the first row considering it has the names of the variables.
How to Add Rows to a Data Frame
To add a row to a data frame, you lot need to apply the rbind function:

This function takes two arguments:
- The data frame that you want to change.
- A list with the data of the new row. To create the listing, yous tin use the
list()role with each value separated past a comma.
This is an example:
> rbind(students_data, list("William", "Smith", xiv, 7, three, "Chocolate-brown")) The output is:
first_name last_name age num_siblings num_pets eye_color one Emily Dawson 15 2 5 BLUE 2 Rose Patterson 14 5 0 GREEN iii Alexander Smith 16 0 2 BROWN four Nora Navona 16 4 ten BROWN 5 Gino Sand 17 iii 8 Blueish half dozen <NA> Smith 14 vii iii BROWN Just wait! A warning message was displayed:
Warning message: In `[<-.cistron`(`*tmp*`, ri, value = "William") : invalid factor level, NA generated And notice the first value of the 6th row, information technology is <NA>:
6 <NA> Smith xiv 7 3 Dark-brown This occurred because the variable first_name was defined automatically as a factor when we read the CSV file and factors take fixed "categories" (levels).
You lot cannot add together a new level (value - "William") to this variable unless you read the CSV file with the value Imitation for the parameter stringsAsFactors, equally shown below:
> students_data <- read.csv("students_data.csv", stringsAsFactors = Imitation) 
At present, if we try to add this row, the data frame is modified successfully.
> students_data <- rbind(students_data, list("William", "Smith", 14, vii, three, "BROWN")) > students_data first_name last_name age num_siblings num_pets eye_color 1 Emily Dawson xv two v Bluish 2 Rose Patterson xiv 5 0 Light-green three Alexander Smith 16 0 2 BROWN 4 Nora Navona 16 iv 10 GREEN 5 Gino Sand 17 3 8 Bluish half dozen William Smith 14 vii three Brown 💡 Tip: Notation that if you read the CSV file again and assign it to the same variable, all the changes fabricated previously volition be removed and you will see the original information frame. Y'all need to add this argument to the first line of code that reads the CSV file and then brand changes to information technology.
How to Add together Columns to a Data Frame
Adding columns to a data frame is much simpler. You need to use this syntax:
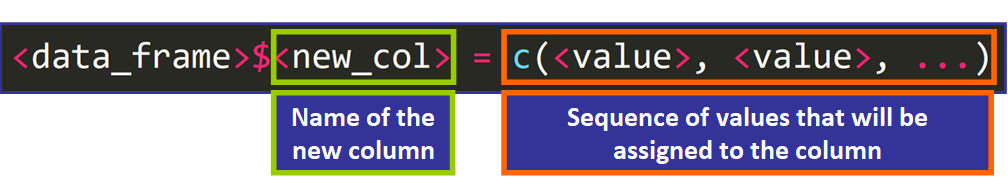
For example:
> students_data$GPA <- c(4.0, 3.5, three.2, iii.15, 2.9, iii.0) 💡 Tip: The number of elements has to be equal to the number of rows of the information frame.
The output shows the data frame with the new GPA cavalcade:
> students_data first_name last_name age num_siblings num_pets eye_color GPA 1 Emily Dawson 15 ii five BLUE 4.00 2 Rose Patterson 14 5 0 GREEN 3.50 3 Alexander Smith 16 0 2 BROWN 3.twenty 4 Nora Navona 16 4 10 GREEN 3.15 5 Gino Sand 17 three eight Blue 2.90 6 William Smith 14 vii 3 BROWN three.00 How to Remove Columns
To remove columns from a data frame, you lot need to utilize this syntax:

When y'all assign the value Null to a cavalcade, that cavalcade is removed from the data frame automatically.
For example, to remove the historic period cavalcade, we use:
> students_data$age <- NULL The output is:
> students_data first_name last_name num_siblings num_pets eye_color GPA 1 Emily Dawson 2 five BLUE 4.00 2 Rose Patterson five 0 GREEN 3.fifty three Alexander Smith 0 two BROWN 3.20 4 Nora Navona 4 10 Green 3.15 five Gino Sand 3 eight BLUE 2.90 half-dozen William Smith 7 iii Chocolate-brown 3.00 How to Remove Rows
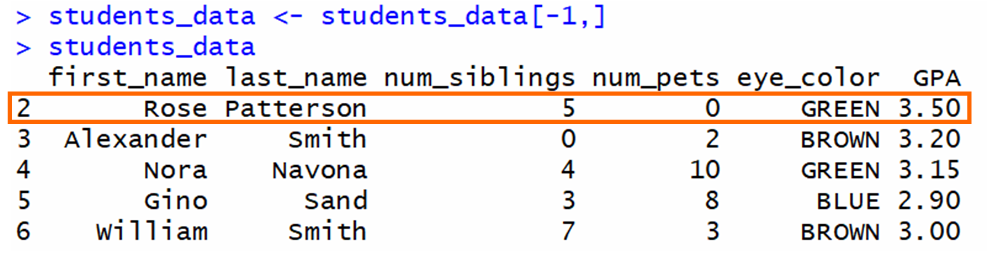
To remove rows from a data frame, you tin utilize indices and ranges. For instance, to remove the first row of a data frame:

The [-one,] takes a portion of the data frame that doesn't include the first row. Then, this portion is assigned to the same variable.
If we have this data frame and we desire to delete the first row:

The output is a data frame that doesn't include the beginning row:
In general, to remove a specific row, you need to utilise this syntax where <row_num> is the row that you want to remove:

💡 Tip: Notice the - sign before the row number.
For example, if we desire to remove row iv from this data frame:

The output is:
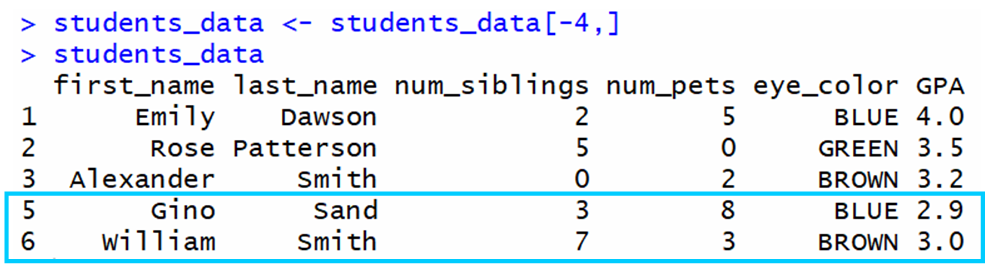
As you can see, row 4 was successfully removed.
🔹 In Summary
- CSV files are Comma-Separated Values Files used to correspond data in the course of a table. These files can be read using R and RStudio.
- Data frames are used in R to represent tabular data. When yous read a CSV file, a information frame is created to shop the data.
- Y'all can access and modify the values, rows, and columns of a data frame.
I really hope that you liked my article and constitute it helpful. At present you tin can work with information frames and CSV files in R.
If you liked this article, consider enrolling in my new online course "Introduction to Statistics in R - A Applied Approach "
Learn to code for free. freeCodeCamp'due south open up source curriculum has helped more forty,000 people become jobs every bit developers. Become started
Source: https://www.freecodecamp.org/news/how-to-work-with-data-frames-and-csv-files-in-r/
0 Response to "Using Rbind for Reading Files With Id"
Post a Comment