How Do Abiotic Factors Effect Marine Feeding Guilds
Abstract
Analysis of environmental DNA (eDNA) offers an unprecedented ability to accurately survey biodiversity from aquatic ecosystems. Although eDNA methods have been applied to myriad taxa, scientists are now moving away from proof-of-concept work, ultimately evaluating the limits and opportunities of this technology to detect and quantify abundance across organisms and environments. Important considerations enabling such methodology to be used for aquatic conservation contexts includes understanding both the effects of (1) the amount of eDNA released from focal taxa—sources, and (2) the removal of eDNA in the environment—sinks. I review publications on aquatic macroorganism eDNA that have evaluated or considered the effect of sources on signal detection (or quantification) and find few studies acknowledge, and fewer still evaluate, the impact of eDNA production on genomic signal recovery. In this review, I encourage readers to carefully consider source dynamics, and using previously published literature, dissect what roles biotic (e.g. life-history traits, species interactions including stressors) and abiotic (e.g. temperature, salinity) factors likely play in eDNA deposition and recovery, and how this impacts detection, abundance, biomass estimation, and ultimately informed signal interpretation. I further explore the physical sources of eDNA and propose other methods (spatial and temporal) and markers to assist in identifying eDNA origins in aquatic systems. Understanding how these parameters influence variation in eDNA sources will allow for a more comprehensive survey tool, and potentially give insights into environment-population responses.
Introduction
Biodiversity assessments are usually the first stage of research associated with natural systems, thus underlying important and varied disciplines including biogeography, restoration ecology, conservation biology, and environmental management and policy (Margurran 2004). Thorough biodiversity surveys however, are often prohibitive, due largely to complications in data collection for hard-to-study taxa (e.g. cryptic, behaviourally elusive, low site fidelity, or rare), difficult to sample locales (e.g. aquatic ecosystems), and affiliated costs. Recent molecular advances now offer an invaluable opportunity to significantly improve the evaluation of previously difficult to attain biodiversity data, making them important tools for aquatic conservation. Yet, our knowledge and use of such molecular sampling techniques still requires refinement.
Environmental DNA (eDNA) is the collective term for genetic material obtained directly from environmental samples (e.g. water, soil, air) without capturing or sampling a target organism. These samples encompass a mixture of genomic DNA released from living or dead organisms within a locale, deposited from diverse sources such as sloughed cells, gametes, metabolic waste, and carcasses (Bohmann et al. 2014). Since eDNA's seminal arrival in macroorganismal surveying in 2008 (Ficetola et al. 2008), it's efficiency, accuracy, and non-invasive application made it an attractive and growing technology, especially for conservation management. Indeed, it is a featured theme for global conservation horizon scans that highlight innovative research opportunities for pragmatic environmental and conservation goals (Sutherland et al. 2013), and is now even being used to assess the ecological status of European waterways (Hering et al. 2018).
To date, studies using eDNA have successfully detected numerous taxa, including fish (Jerde et al. 2011; Takahara et al. 2012), amphibians (Dejean et al. 2012; Pilliod et al. 2013; Harper et al. 2018), reptiles (Hunter et al. 2015), mammals (Foote et al. 2012; Ma et al. 2016; Stewart et al. 2017), invertebrates (Goldberg et al. 2013; Deiner and Altermatt 2014), and aquatic plants (Scriver et al. 2015; Fujiwara et al. 2016), both in the lab and field. This transformation to biodiversity assessments can in part be attributed to eDNA's ability to accurately survey large portions of the living environment in a cost- and time-effective manner, offering increased spatial and temporal resolution (e.g. Bista et al. 2017) when compared to traditional/manual methods of environmental sampling (Davy et al. 2015). Although extremely promising, scientists are still circumscribing the saliency of this technology and have started to evolve away from preliminary studies that provide proof-of-concept information regarding presence/absence, to more nuanced investigations surrounding abundances. These include the development of theoretical expectations and empirical testing, thus ultimately evaluating eDNA's potential and limitations across taxa and environments.
Despite the revolution eDNA has afforded aquatic conservation scientists and managers, there are two important variables known to impact eDNA abundance estimations that have not, as yet, received equal consideration. The concentration of eDNA in the aquatic environment remains highly variable as a product of (1) the amount of eDNA released from focal taxa—sources, and (2) the removal of eDNA in the environment—sinks. A large proportion of the published literature thus far has been devoted to understanding how sinks affect eDNA detection and quantification, including its persistence and degradation (Dejean et al. 2011; Barnes et al. 2014; Merkes et al. 2014; Piaggio et al. 2014; Pilliod et al. 2014; Ma et al. 2016), transportation (Deiner and Altermatt 2014; Jane et al. 2015), and location and settlement (Moyer et al. 2014; Turner et al. 2015) in the environment. Although a few studies have attempted to quantify the amount of eDNA released by an organism over time (e.g. Thomsen et al. 2012a; Pilliod et al. 2014; Klymus et al. 2015), in general less consideration has been given to the sources (i.e. production rate) of eDNA and how it might influence detection, quantification, and data interpretation. Understanding associations between species abundance and amplicon abundance (Doi et al. 2016; Evans et al. 2016; Kelly 2016), and elucidating the influences of abiotic and biotic parameters on eDNA production, are important research goals for interpreting these data in a conservation context.
In this review, I examine peer-reviewed publications to determine the number of studies that have analyzed production rate/sources of eDNA. I then discuss factors that are likely to impact the production of aquatic eDNA, physical sources of this genomic material, and why understanding sources, and how best to approach abundance estimation, are important next-steps for research utilizing eDNA technology for aquatic biodiversity assessments and conservation applications.
Literature review
I searched The Web of Science for peer-reviewed journal papers using keywords 'environmental DNA' and 'eDNA', restricting the review to aquatic studies and macroorganisms (both representing the current predominant foci in conservation practices; sensu Dudgeon et al. 2006). As eDNA methods to date have not been extensively used to monitor aquatic plants (but see Scriver et al. 2015; Fujiwara et al. 2016), and because plants may not always release detectable amounts of DNA (e.g. Matsuhashi et al. 2016) suggesting the use of eDNA methods for plant conservation may still be in its infancy, I only concentrate on animal studies herein (but recognise some key points may be transferable).
The literature search was conducted on 7th November 2016 and covered the years 2008 to the search date. This investigation yielded 170 publications (Appendix S1). A total of 27 studies systematically evaluated sources of eDNA (including production rate, shedding, excretion, deposition), with 22 looking at biotic factors and 7 abiotic factors (2 investigated both biotic and abiotic; Table 1), accounting for only 15.9% of the total eDNA papers published to date. Although not investigated specifically, 41 papers acknowledged source factors (i.e. variation in production rate) as a potential influence on eDNA detection or abundance (Appendix S2), accounting for a further 24.1%. The remaining 60% of publications included those evaluating eDNA sinks, transportation or settlement, or review papers, none of which discuss eDNA sources.
The axiom of investigating sources of eDNA is fundamentally linked to the focal organisms under investigation, and incorporating species-specific life-history and ecology has a long history of being fraught with dynamic or coupled variables that are often in flux with the environment. As it is, we know very little about the ecology of many organisms, and unsurprisingly, a disproportionate amount of eDNA research focuses on a few, relatively well-studied species (e.g. common carp, Cypinus carpio: Takahara et al. 2012; Barnes et al. 2014; Eichmiller et al. 2014; Turner et al. 2014; American bullfrog, Lithobates catesbeianus: Dejean et al. 2011; Stickler et al. 2015; Evans et al. 2016). Although much could be said about expanding our knowledge regarding how eDNA is produced or removed across myriad species and ecosystems, these well-studied model systems may lend invaluable opportunities to gain a more in-depth understanding on eDNA source dynamics.
Given recent evidence for idiosyncratic eDNA production, trepidation about the utility of eDNA as a surveying tool has recently been raised (e.g. Iversten and Kielgast 2015). Certainly abundant research to date exemplifies the value of genomic over various traditional methodologies for biodiversity surveys, yet examining source/sink eDNA dynamics warrants careful consideration prior to implementation. Below I explore factors that conceivably impact eDNA sources (Fig. 1), and where possible, lead scientists toward avenues of research that merit evaluation or deliberation when employing these tools in an aquatic conservation context.
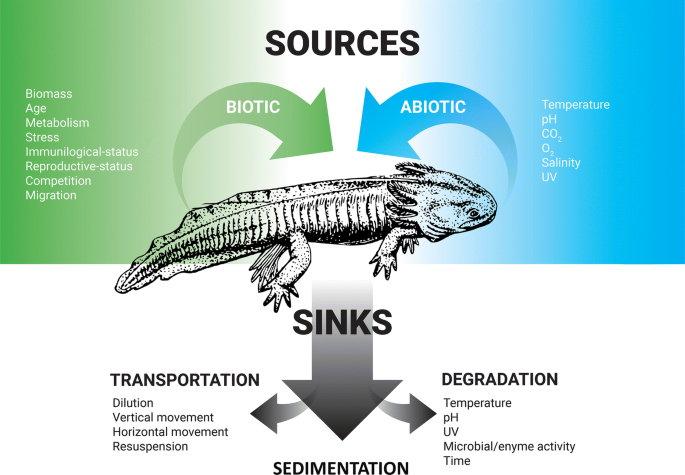
Summary diagram illustrating the known associations of various eDNA sources (biotic and abiotic) and sinks that may affect detection and abundance quantification for aquatic macrofauna
Biotic factors
Life-history
Knowledge about the life-history of focal taxa is desirable but can be unknown when designing eDNA sampling strategies. Still, understanding associations between an organism's size, age, condition, or biological activity and the production of DNA within the environment will assist in determining accurate relationships between eDNA concentration and relative abundance measures. These species and habitat-specific nuances highlight the importance of a priori organismal biology and phenological pattern knowledge.
While seasonality-eDNA studies still remain scant in the literature (but see Stoeckle et al. 2017; Bista et al. 2017), those that have considered this attribute often find marked increases in the abundance of eDNA signals during breeding seasons. For example, eDNA signals have been shown to spike during the spawning of fish [e.g. Oriental weatherloach, Misgurnus anguillicaudatus (Lintermans et al. 2007), Chinook Salmon, Oncorhychnus tshawytscha (Laramie et al. 2015), and invasive Bigheaded Carp, Hypophthalmichthys spp. (Erickson et al. 2016)]. Similarly, strong seasonal influences to eDNA detection have also been seen in both amphibians and reptiles, presumably as a reflection of species-specific organismal behaviour (de Sousa et al. de Souza et al. 2016) such as male–male combat and the mass release of gametes (e.g. Eastern Hellbender, Cryptobranchus alleganiensis (Spear et al. 2015)). Genetic material such as gametes, blood, and other reproductive tissues (e.g. placenta, lactation), all combine to make breeding events optimal for the detection of eDNA in wild populations. However, in additional to giving us windows into breeding behaviour and geographic locations that may have been previously obscured (e.g. Stewart et al. 2017; Stoeckle et al. 2017), it may also over-estimate the relative presence of a species both temporally and spatially. Understanding when and where reproductive bouts occur in a species, and comparing eDNA detection and abundance quantification during such events to non-breeding times, will greatly assist in our understanding of how these genomic sources affect biomass predictions.
Species-specific differences may also influence the quantity and source of eDNA production, strongly influenced by not only the size but also ecology of target taxa. Indeed, studies have reported disparities in eDNA sources across and within taxonomic groups (e.g. Goldberg et al. 2011; Thomsen et al. 2012a; Sassoubre et al. 2016). For example, eDNA detection has been shown to vary between Idaho giant salamanders (Dicamptodon aterrimus) and Rocky Mountain tailed frogs (Ascaphus montanus) in the same stream (Goldberg et al. 2011), suggesting species disparities are not merely a consequence of environmental effects. The production rates of eDNA were also found to differ by Thomsen et al. (2012a) between two juvenile amphibian species, two fish species (2012b), and also by Sassoubre et al. (2016) among three fish species, all stemming from mesocosms experiments. Detection differences were observed across six macroinvertebrate species whereby a single species (Tinodes waeneri) was not detected despite its presence (Mächler et al. 2014). In this case, it was suggested that T. waeneri (a caddisfly with a sand case) may have limited or delayed release of DNA due to its distinct life-history (Mächler et al. 2014).
Evidence from real-time quantitative PCR also indicates eDNA concentration is positively correlated to individual biomass (Takahara et al. 2012; Pilliod et al. 2014; Piggott 2016). However, these studies have estimated fish biomass using linear or exponential regressions which do not model abiotic and biotic parameters (Sassoubre et al. 2016). Read abundance has additionally been positively associated with biomass abundance via metabarcoding, although high variability within controlled mesocosm experiments (albeit with weak positive associations) have raised understandable concern over whether these relationships can be translated into more complex natural systems (e.g. Evans et al. 2016). In point of fact, collecting samples from mesocosm experiments can endeavour to parse-out the relative contributions of eDNA sources over sinks, however metabolic rates in stable lab experiments often overestimate metabolic activity (e.g. by up to 35% compared to free-living populations; Auer et al. 2016), thus researchers should be cautious about drawing linear comparisons between the two systems. Size differences between species (biological differences) and within a species (e.g. age structure or morphometry) are likely to influence eDNA production and biomass interpretations, and thus incorporating these covariates into models will likely render eDNA inferences more reliable.
Consideration should also be given to the age-structure of wild populations as eDNA detection differences due to life-history stage have been recurrently observed. For example, incongruences in eDNA production between juveniles and adults has been demonstrated in fish (Maruyama et al. 2014), amphibians (Goldberg et al. 2011; Thomsen et al. 2012a), and invertebrates (Treguier et al. 2014). In aquaria experiments on bluegill sunfish (Lepomis macrochirus) for instance, Maruyama et al. (2014) found that despite eDNA sources being positively related to biomass, juveniles had a slightly higher excretion rate compared to adults, possibly due to adult ontogenetic reduction in metabolic activity (Brett and Groves 1979). Moreover, higher eDNA detection for juvenile crayfish (Procambarus clarkii) was posited to be a result of increased moulting in early stages of growth (Treguier et al. 2014) as exuviae are a known source of DNA (Watts et al. 2005). The risk in using eDNA abundance measures in wild populations without the consideration of age-structure would be incorrect population abundance estimations, especially if populations are dominated by one age class or another (Maruyama et al. 2014) such as in dwindling populations with low birth rates, or newly founded populations monopolised by juveniles.
Species interactions
How organisms respond in the presence of others is another plausible biotic factor affecting eDNA production that needs careful attention. Empirical evidence suggests higher eDNA abundance simply reflect higher population density (Thomsen et al. 2012a; Pilliod et al. 2014; Klymus et al. 2015), yet rarely in complex environments does one find populations consisting solely of an individual species. Sassoubre et al. (2016) observed eDNA production rates of mackerel (Scomber japonicas), anchovy (Engraulis mordax), and sardines (Sardinops sagax) were not affected by the presence of the other individuals of the same species (on a per gram basis) but there were significant increases in eDNA production when different species were housed together. Additionally, under certain conditions or primers, high abundance species have been shown to 'mask' the detection of low abundance species (Erickson et al. 2016; Evans et al. 2016). The impact of predator–prey interactions on eDNA production rates is to date unknown, but physiological impacts of predators on prey include reduced food intake, increased metabolic rate, and increased stress (Thaler et al. 2012; Boonstra 2013; Van Dievel et al. 2016), suggesting ensuing alterations to eDNA sources.
Physiological factors are assumed to form close associations between metabolic and excretion rates, the latter of which is the expected major physical source of eDNA in aquatic systems. Acute perturbations to physiological homeostasis, such as stress, have been indicated as having possible effects on sources of genomic material (Pilliod et al. 2014; Klymus et al. 2015). Increased eDNA production, for instance, was observed in several studies due to osmotic changes or following animal handling (Takahara et al. 2012; Maruyama et al. 2014; Pilliod et al. 2014; Sassoubre et al. 2016). This early spike in eDNA concentration following the introduction of a focal species into a new environment has likewise been observed in an invasive aquatic plant (Egeria densa; Fujiwara et al. 2016) and could suggest a salient response of genomic deposition across animal and plant taxa. Furthermore, inconsistent genomic production has been observed in animals infected with disease, such as before mass die-offs in amphibian populations suffering from ranavirus (Hall et al. 2016). Increases in density (McKenzie et al. 2012), social tension (Sloman et al. 2000) and shelter availability (Milidine et al. 1995), are further examples of stress that affect metabolic activity in aquatic organisms with potential feedbacks on eDNA sources, warranting further investigation.
In addition to metabolic changes, stress may also activate immune responses in skin, especially from illness and parasites. Skin-mucus properties have been shown to shift in common carp (C. carpio) in response to increases in overall bacterial load in the water (Van der Marel et al. 2010), and amoebic gill disease can also cause a whole-body mucus response in some fish species (Roberts and Powell 2005). Immune responses that result in an increase in mucus and epithelial cell turnover predictably increase eDNA excretion in many species. Moreover, parasites and disease have been shown to be energetically demanding in fish (Gomez et al. 2016), and affect feeding behaviour in tadpoles (DeMarchi et al. 2015). Still, in many systems the cost(s) of parasitic infection and disease on host physiology is unknown, but these acute and chronic responses need consideration when using eDNA to quantify abundance.
Abiotic factors
Temperature and salinity
Certainly most aquatic animals specialise within a narrow range of abiotic factors that results in trade-offs at several hierarchical levels, from molecular structure to whole-organism functioning and behaviour (Pörtner 2010). In ectotherms for example, simple maintenance-metabolism reacts as a function of temperature (Tirsgaard et al. 2015).
Temperature, and concomitant microbial activity, has been demonstrated to increase DNA release from plant matter within aquatic sediments (Poté et al. 2009) and these patterns may also be reflected in animal taxa. Indeed, numerous studies suggest metabolism, growth, physiology, and immune function in fish are all influenced by water temperature (Engelsma et al. 2003; Person-Le Ruyet et al. 2004; Takahara et al. 2011), which in-turn may increase the excretion of mucus and shedding of epithelial cells of aquatic macrofauna. As a by-product of metabolic influences, evidence suggests temperature further affects the production of faeces and urine in fish (Selong et al. 2001; Gale et al. 2013), presumably the primary component of eDNA sources. Fish mobility is increased with water temperature (Petty et al. 2012) and thus genetic signals may additionally be more homogenised and/or spatially dispersed. To date, three mesocosm studies specifically examining effects of temperature on eDNA production rates have found conflicting results; no effect in two studies (common carp, C. carpio, Takahara et al. 2012; bighead carp, Hypophthalmichthys spp., Klymus et al. 2014), and a significant increase in production rates in Mozambique tilapia (Oreochromis mossambicus; Robson et al. 2016). In a field study, high water temperature significantly increased the amount of Brook Charr (Salvelinus fontinalis) eDNA within the water column, and moreover, biomass and thus population abundance predictability increased in higher temperatures (Lacoursiere-Roussel et al. 2016). Field collections have also seen higher eDNA concentrations in pools with warmer compared to cooler water, although this may have resulted from organismal attraction, and thus a resultant increase in population size, rather than an effect of temperature on eDNA sources per se (Takahara et al. 2012). Large temperature ranges among and within lakes, especially in temperate regions where seasonal, longitudinal, and latitudinal variations can impose substantial impacts on eDNA concentrations, should consequently be incorporated into predictive models in natural systems (Lacoursiere-Roussel et al. 2016).
Similar to biotic seasonal effects on eDNA production such as breeding behaviour, temperature itself can impact the excretion of genetic material into the environment when phenologies concurrently affect other organismal physiological (e.g. metabolic regulation) or behavioural responses (e.g. temporal avoidance). For example, although eDNA has yet to be specifically quantified, overwintering salmonid fry (Oncorhynchus spp.) demonstrate very low standard metabolic rates as a direct response to low temperatures (Eliason and Farrell, 2016). More specifically, eDNA detection of Idaho giant salamanders (D. aterrimus) and Rocky Mountain tailed frogs (A. montanus) was observed to be lower in early spring (Goldberg et al. 2011), ostensibly due to a combination of decreased metabolism or moving into the hyporheic zone. Daily migration behaviour such as diel vertical migration (e.g. Levy 1990; Armstrong et al. 2013) further impacts eDNA detection depending on where species are in the water column (Stewart et al. 2017), DNA persistence, and time of sampling.
Exceedingly, researchers speculate that DNA location within the environment reflects preservation or decomposition rates (Moyer et al. 2014; Turner et al. 2015). However, source dynamics are also plausible contributors to DNA location, wherein sites with high eDNA concentrations may elucidate organismal behaviour and ecosystem characteristics. To passively avoid temperature limits that may induce heat-shock, some intertidal taxa employ vertical zonation for example (Somero 2002), a behavioural response that should be considered when interpreting eDNA patterns. Certainly, coarse spatio-temporal fluctuations in water temperature, and to a lesser extent fine-scale idiosyncrasies, likely have downstream effects on eDNA production and thus our inferences as to population biomass. Populations of the same species can also undoubtedly vary in habitat or phenotype (e.g. ecomorphs) where the potential for producing genetic material at vastly dissimilar rates even within different thermal habitats of the same water body may be high.
In a similar manner as temperature responses, adaptation to saline environments also requires physiological compensation and acclimation. Egg fertilization and incubation, early embryogenesis, swim bladder inflation, and larval growth in most fish species are all dependent on salinity (Boeuf and Payan 2001). In fact, studies have shown that up to 50% of fishes total energy budget may be dedicated to osmoregulation (Bushnell and Brill 1992), with food intake, food conversion, and hormones associated with growth regulation dependent on environmental salinity (Boeuf and Payan 2001). Smoltification in salmon, for instance, has demonstrated drastic physiological adjustment to saltwater, with significantly different metabolism to that of their parr freshwater counterparts (e.g. McCormick et al. 1989). The pervasive links between salinity and fish growth has been shown for both marine and freshwater species, with general patterns suggesting marine species growth rates are increased in slightly lower saline environments, whereas freshwater species development show the opposite relationship (Boeuf and Payan 2001). Granted, assessing marine species richness and approximate abundance is a relatively new foray for eDNA (e.g.Günther et al. 2018; Knudsen et al. 2019) and has proven successful for accurate detection, read abundance has failed to find correlations with DNA proportions (Günther et al. 2018) or traditional visual biomass measures such as trawling (Knudsen et al. 2019). Undoubtedly marine and freshwater systems are likely to experience difference abiotic parameters affecting eDNA dynamics, but whether production rates in marine systems vary in vastly dissimilar ways compared to freshwater habitats, is a yet unknown facet of this methodology.
Other influences
Abiotic parameters for aquatic macrofauna can ultimately act directly through receptors to increase/decrease growth (e.g. temperature, salinity) or can be a limiting threshold within a tolerance range (e.g. pH, CO2, O2). Often acting synergistically (e.g. with temperature), pH, UV radiation, CO2, and O2 have complex interactions, and decoupling how these components influence eDNA sources may be difficult. Increasing temperature for example, decreases saturation concentrations of O2 due to the decreased capacity of water to carry oxygen, and aerobic performance is often limited by high temperatures (Bozinovic and Pörtner 2015) as seen in crustaceans (e.g. Storch et al. 2009) and zooplankton (Seidl et al. 2005). In aquatic ecosystems with fluctuating O2 levels (e.g. lakes or streams), dissolved O2 depletion could increase the frequency of hypoxia status. Reduced spawning, sperm motility, fertilization success, hatching rate, and larval survival have been described as hypoxia effects on wild fish populations (Wu et al. 2003), which although only speculative, likely have large impacts on eDNA sources at a population scale.
Confluences in various abiotic parameters further exist, for instance, intertidal fish increase O2 consumption and demonstrate a disruption to body growth when exposed to high UV radiation, revealing the power of UV on respiration and energy expenditure in fish (García-Huidobro et al. 2017). Increasing temperatures and concomitant summer droughts favour acidic environmental conditions in surface waters due to higher CO2 production (Wright 2008). Rates of survival, reproduction, hatching success, swimming behaviour, and body chemistry of both fish and aquatic invertebrate species (Jordahl and Benson 1987; Havas and Rosseland 1995) have all been shown to be affected by pH, with early developmental stages being more sensitive to pH variations. In amphibians specifically, embryonic development and growth curves are stunted in acidic environments, and evidence also demonstrates reduced tadpole swimming performance (Arena-Rodríguez et al. 2016). What's more, changes in aquatic pH may also affect the biotic community and translate into alterations in population structure or reduced species diversity.
Of the limited eDNA research that has been done on these abiotic influences to date, Seymour et al. (2018) demonstrate eDNA quantification to be 1–2 orders of magnitude greater in basic compared to acidic sites. However, whether this solely results from differential degradation rates (e.g. Strickler et al. 2015) rather than DNA production has yet to be determined. On the other hand, Buxton et al. (2017) found pH to be of little influence on detection rates in mesocosms. Certainly, systematic research quantifying how these abiotic parameters influence the production of DNA being released from focal organisms (paralleling analysis of abiotic influences on eDNA degradation rates; Barnes et al. 2014) will expand the utility of aquatic eDNA tools for conservation purposes, especially in light of rapidly changing environments (e.g. climate change and species range shifts).
Finally, it's important to note that input of genetic material may further become redistributed within aquatic systems via abiotic processes such as flow/discharge (e.g. Deiner and Altermatt 2014; Jane et al. 2015) or from previous particle settlement (e.g. sedimentation; Turner et al. 2015). While these signals are often described as sinks of eDNA for their propensity to remove DNA from their place of origin (Fig. 1), they could also be argued sources of eDNA, despite not currently being produced by focal organisms at a given location. Still, these abiotic processes have been shown to render longer-lasting and abundant genetic signals (Turner et al. 2015) from locations far from their origin (Deiner and Altermatt 2014), thus I advocate practitioners using eDNA for conservation should be aware of their influence on eDNA detection/quantification.
How best to approach abundance estimation?
Given what we know about biotic and abiotic factors affecting eDNA production, I propose several avenues of research below to help improve eDNA abundance estimation in aquatic environments for conservation purposes: i. identifying the physical origins of eDNA and ii. modifying experimental design, especially sampling, for the retrieval of these data.
Physical origins
To date, research on eDNA physical origins has been limited to particle size distribution (Turner et al. 2014). Although Turner et al. (2014) found particle size most likely to correspond to individual cells or mitochondria, the physical identity of eDNA still remains largely unknown. Based on earlier work, faecal origins from focal species are assumed the most likely sources of eDNA in aquatic environments (Martellini et al. 2005), presumably entering the water column in large particles, thereafter rapidly settling or breaking apart (Turner et al. 2014). Faeces from aquatic macrofauna are known to rapidly sink (Robison and Bailey 1981; Wotton and Malmqvist 2001) suggesting that even if faeces are a large source of eDNA, this source may not be available for long in the water column. Supported by evidence of concentrated eDNA in sediments compared to water samples (Turner et al. 2015), faecal origins seem the most prominent of eDNA sources, but exceptions to this are those species whose faeces are known to float, such as marine mammals (Parsons et al. 1999; Carney et al. 2007; Gillett et al. 2008).
Predator faeces from outside the principal system (allochthonous eDNA; Goldberg et al. 2016) however, may also contribute to DNA signals found within a habitat, although the magnitude and persistence of its input remains uninvestigated. Studies on amphibians (Ficetola et al. 2008) and fish (Jerde et al. 2011) further suggest bodily fluids such as slime coatings can act as primary sources for eDNA (Ficetola et al. 2008; Jerde et al. 2011; Pilliod et al. 2014), although Merkes et al. (2014) found slime a more variable source of DNA compared to carcasses. Even among similar taxa, there are likely to be differences in excretion, such as scales versus mucus in fish species (Sassoubre et al. 2016). While the presence of an exoskeleton in aquatic arthropods may reduce extracellular DNA release (Treguier et al. 2014), for some taxa, exoskeletons are shed at known times of the year generating sharp increases in eDNA production. Data also demonstrate spawning activities increase eDNA and thus organismal detection (e.g. Gustavson et al. 2015; Spear et al. 2015; Erickson et al. 2016), signifying gametes and other reproductive-related fluids and tissues likely compose a large proportion of eDNA sources, at least seasonally. A final, albeit unusual and likely small potential eDNA source contributor, is exhaled breath condensate from whales and dolphins (Frère et al. 2010). Despite its importance for eDNA and population interpretation, distinguishing among these eDNA physical sources has proven difficult to date.
Identifying the physical sources of eDNA without in-depth taxonomic expertise may still be tractable, and further help to infer production rate physiological foundations. RNA (a molecule with lower stability than DNA, and thus higher temporal sensitivity) has, for example, been utilised to identify various bodily fluids at crime scenes (Vennemann and Koppelkamm 2010). In fact, mRNA forensic studies have demonstrated feasibility at detecting and distinguishing between skin, saliva, blood, and reproductive fluids (Haas et al. 2009; Hanson and Ballantyne 2014; Nakanishi et al. 2017) even in old, degraded samples (Zubakov et al. 2009). Although only recently investigated, eRNA has shown promise at distinguishing living biodiversity with metabolically active microorganisms, to potential legacy DNA signals (e.g. Capo et al. 2015; Pochon et al. 2017). Where eRNA's short persistence time makes it a unique tool to distinguish dead from living organisms, it's rapid degradation may make it equally challenging to work with. Ultimately, whether similar patterns of eDNA/eRNA detection translate to work involving macroorganisms is unclear. Additionally, although nuclear fragments within environmental samples may be less abundant than mitochondrial (Bylemans et al. 2018), nuclear markers may be more sensitive for eDNA detection (Minamoto et al. 2017), and in addition to extending the potentiality of eDNA data to address questions in population genetics and introgression (Piggott 2016; Minamoto et al. 2017), nuclear markers can successfully detect gametes and spawning activity in wild populations (Bylemans et al. 2016). Transcriptomics and identifying protein expression profiles may further provide clues toward stimuli responses, organismal activity, or ecosystem health (Barnes and Turner 2016), an avenue of study already successfully navigated in microbial research (Maron et al. 2007). Foote et al. (2012) further suggest differentiating between cellular and extracellular DNA, as the latter degrades faster with a higher propensity to demonstrate living organisms (sources) rather than degradation or transported genomic material (sinks). Finally, as suggested by Barnes and Turner (2016), microscopy-based investigations, such as delineating faecal from gamete or carcase inputs, may be of use to determine the origins of eDNA. These methodological compliments to eDNA research should further dissect the type of genomic material being collected, and thus inferences on individual and population health, a cornerstone to impactful conservation work.
Experimental design
Alternatively, consideration of spatial and temporal sampling may assist in discerning eDNA sources and further support more accurate abundance estimations. One means to circumnavigate the issue of living and dead sources of eDNA is identifying temporal fluctuations in genomic signals through multiple sampling events. Species actively present within a system should illustrate continuous detection compared with carcasses that decrease eDNA release over time (e.g. Dunker et al. 2016). Temporal fluctuations to eDNA production however, could infer seasonal population fluctuations or be due to intermittent sediment mixing, such as during moments of high turbidity or seasonal turnover of thermally stratified layers. Disentangling these patterns remains arduous and complex, demonstrating the need for more practical means of eDNA differentiation.
Strategic spatial sampling may represent a simpler means to assist in discerning eDNA sources. For instance, unlike fish faeces, marine mammal faeces float (Parsons et al. 1999; Carney et al. 2007; Roman and McCarthy 2010) and upper marine layers can be easily targeted to acquire the eDNA of living whale, manatee, dolphin, and pinniped species. Similarly, fish gametes have also demonstrated species-specific vertical distributions in marine environments (Sundby and Kristiansen 2015) allowing researchers to specifically quantify eDNA from reproductive bouts. However, egg buoyancy and ensuing vertical egg distribution may change during development (Ospina-Álvarez et al. 2012), further highlighting the importance of species-specific natural history knowledge.
Conclusions
Environmental DNA is quickly becoming one of the fastest developing and utilized tools currently employed for non-invasive biodiversity surveillance and quantification. It is also successfully driving rapid advancement in conservation biology, ecology, and evolutionary biology, yet some challenges remain, especially regarding methodological standardization, quantification strategies, and data interpretation. Thus, even minor advancements to the general knowledge of design and application of eDNA tools in natural populations helps to propel the entire field to a more thorough understanding of utility and limitations.
Understanding the mechanistic underpinnings of, and sensitivity to, biotic and abiotic parameters on the production of aquatic eDNA will surely help to facilitate a more comprehensive tool for abundance measures and conservation management. Though individual variance (intra- and interspecific) in DNA production may raise queries as to the applicability of eDNA as a tool for abundance measures (e.g. Iversten and Kielgast 2015), eDNA signals can further give insights into environmental responses, but care is warranted. Biotic and abiotic factors, for example, are not mutually exclusive and their interplay likely have a compounding effect on physiological responses and thus eDNA sources. As practitioners move forward with the use of eDNA protocols, a tool well-suited to conservation goals over numerous traditional survey techniques, critical consideration needs to be given to the sources of these data through either the investigation of biotic and abiotic covariates, eDNA physical sources, or via the incorporation of system- or species-specific temporal and spatial sampling.
Environmental DNA has afforded scientists a means to quantify the distribution and abundance of species, but what information can the genetic residues of individuals inhabiting those ecosystems tell us about their responses to their surroundings? For example, opportunities exist to link eDNA survey data with species distribution modelling (Dorazio and Erickson 2018), comparing and contrasting where eDNA metrics and traditional species abundance estimations are over- or under-represented (e.g. Schmelzle and Kinziger 2018). Advancements to non-invasive genetic monitoring, such as the inclusion of ecologically based species-specific sampling sites (e.g. hydrological zones), and novel genetic markers and methodologies (e.g. eRNA, transcriptomics, and nuclear markers), may help to galvanise eDNA methods in gaining invaluable insights beyond mere presence and abundance measures.
Data accessibility
This manuscript does not include data.
References
-
Arena-Rodríguez A, Beltrán MA, Bernal MH (2016) Lethal and sublethal effects of acidic and basic pH in the embryos and tadpoles of Colombian anurans: acute toxicity under microcosm conditions. Int J Adv Life Sci 9:428–437
-
Armstrong J, Schindler D, Ruff C, Brooks GT, Bentley KE, Torgersen CE (2013) Diel horizontal migration in streams: juvenile fish exploit spatial heterogeneity in thermal and trophic resources. Ecology 94:2066–2075
-
Auer SK, Bassar RD, Salin K, Metcalfe NB (2016) Repeatability of metabolic rate is lower for animals living under field versus laboratory conditions. J Exp Biol 219:631–634
-
Barnes MA, Turner CR (2016) The ecology of environmental DNA and implications for conservation genetics. Conserv Genet 17:1–17
-
Barnes MA, Turner CR, Jerde CL, Renshaw MA, Chadderton WL, Lodge DM (2014) Environmental conditions influence eDNA persistence in aquatic systems. Environ Sci Technol 48:1819–1827
-
Bista I, Carvalho GR, Walsh K, Seymour M, Hajibabaei M, Lallias D et al (2017) Annual time-series analysis of aqueous eDNA reveals ecologically relevant dynamics of lake ecosystem biodiversity. Nat Commun 8:14087
-
Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M et al (2014) Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol 29:485
-
Boonstra R (2013) Reality as the leading cause of stress: rethinking the impact of chronic stress in nature. Funct Ecol 27:11–23
-
Boeuf G, Payan P (2001) How should salinity influence fish growth? Comp Physiol C Toxicol Pharmacol 130(4):411–423
-
Bozinovic F, Pörtner H-O (2015) Physiological ecology meets climate change. Ecol Evol 5:1025–1030
-
Brett J, Groves T (1979) Physiological energetics. In: Hoar W, Brett J (eds) Fish physiology. Academic Press, New York, pp 279–352
-
Bushnell PG, Brill RW (1992) Oxygen transport and cardiovascular responses in skipjack tuna (Katsuwonus pelamis) and yellowfin tuna (Thunnus albacares) exposed to acute hypoxia. J Comp Physiol B 162:131–143
-
Buxton AS, Groombridge JJ, Griffiths RA (2017) Is the detection of aquatic environmental DNA influenced by substrate type. PLoS ONE 12(8):e0183371
-
Bylemans J, Furlan E, Hardy C, McGuffie P, Lintermans M, Gleeson DM (2016) An environmental DNA (eDNA) based method for monitoring spawning activity: a case study using the endangered Macquarie perch (Macquaria australasica). Methods Ecol Evol. https://doi.org/10.1111/2041-210x.12709
-
Bylemans J, Furlan EM, Gleeson DM, Hardy CM, Duncan RP (2018) Does size matter? An experimental evaluation of the relative abundance and decay rates of aquatic environmental DNA. Environ Sci Technol 52(11):6408–6416
-
Capo E, Debroas D, Arnaud F, Domaizon I (2015) Is plaktonic diversity well recorded in sedimentary DNA? Toward the reconstruction of past protistan diversity. Microb Ecol 70(4):865–875
-
Carney SL, Bolen EE, Barton SL, Scolardi KM, Englund CC, Tringali MD, Reynolds JE III (2007) A minimally invasive method of field sampling for genetic analyses of the Florida manatee (Trichechus manatus latirostris). Mar Mamm Sci 23:967–975
-
Davy CM, Kidd AG, Wilson CC (2015) Development and validation of environmental DNA (eDNA) markers for detection of freshwater turtles. PLoS ONE 10:e0130965
-
de Souza LS, Godwin JC, Renshaw MA, Larson E (2016) Environmental DNA (eDNA) detection probability is influenced by seasonal activity of organisms. PLoS ONE 11:e0165273
-
Deiner K, Altermatt F (2014) Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 9:e88786
-
Dejean T, Valentini A, Duparc A, Pellier-Cuit S, Pompanon F, Taberlet P, Miaud C (2011) Persistence of environmental DNA in freshwater ecosystems. PLoS ONE 6:e23398
-
Dejean T, Valentini A, Miquel C, Taberlet P, Bellemain E, Miaud C (2012) Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. J Appl Ecol 49:953–959
-
DeMarchi J, Gaston J, Spadaro A, Porterfield C, Venesky M (2015) Tadpole food consumption decreases with increasing Batrachochytrium dendrobatidis infection intensity. J Herpetol 49:395–398
-
Doi H, Uchii K, Takahara T, Matsuhashi S, Yamanaka H, Minamoto T (2016) Use of droplet digital PCR for estimation of fish abundance and biomass in environmental DNA surveys. PLoS ONE 10(3):e0122763
-
Dorazio RM, Erickson RA (2018) eDNAoccupancy: An R package for multiscale occupancy modelling of environmental DNA data. Mol Ecol Resour 18(2):368–380
-
Dudgeon D, Arthington AH, Gessner MO, Kawabata Z-I, Knowler DJ et al (2006) Freshwater biodiversity: importance, threats, status and conservation challenges. Biol Rev 81:163–182
-
Dunker KJ, Sepulveda AJ, Massengill RL, Olsen JB, Russ OL, Wenburg JK, Antonovich A (2016) Potential of environmental DNA to evaluate northern pike (Esox lucius) eradication efforts: An experimental test and case study. PLoS ONE 11:e0162277
-
Eichmiller JJ, Bajer PG, Sorenson PW (2014) The relationship between the distribution of common carp and their environmental DNA in a small lake. PLoS ONE 9:e112611
-
Eliason E, Farrell A (2016) Oxygen uptake in Pacific salmon Oncorhynchus spp.: when ecology and physiology meet. J Fish Biol 88:359–388
-
Engelsma M, Hougee S, Nap D, Hofenk M, Rombout JHWM, van Miuswinkel WB, Lidy Verburg-van BM (2003) Multiple acute temperature stress affects leucocyte populations and antibody responses in common carp, Cyprinus carpio L. Fish Shellfish Immunol 15:397–410
-
Erickson RA, Rees CB, Coulter AA, Merkes CM, McCalla SG, Touzinsky KF et al (2016) Detecting the movement and spawning activity of bigheaded carps with environmental DNA. Mol Ecol Resour 16:957–965
-
Evans N, Olds BP, Renshaw MA, Turner C, Li Y, Jerde C et al (2016) Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Mol Ecol Resour 16:25–41
-
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4:423–425
-
Foote AD, Thomsen PF, Sveegaard S, Wahlberg M, Kielgast J, Kyhn LA et al (2012) Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE 7:e41781
-
Frère CH, Krzyszczyk E, Patterson EM, Hunter S, Ginsburg A, Mann J (2010) Thar she blows! A novel method for DNA collection from cetacean blow. PLoS ONE 5:e12299
-
Fujiwara A, Matsuhashi S, Doi H, Yamamoto S, Minamoto T (2016) Use of environmental DNA to survey the distribution of an invasive submerged plant in ponds. Freshw Sci 35:748–754
-
Gale MK, Hinch SG, Donaldson MR (2013) The role of temperature in the capture and release of fish. Fish Fish 14:1–33
-
García-Huidobro R, Aldana M, Duarte C, Galbán-Malagón C, Pulgar J (2017) Seawater-temperature and UV-radiation interaction modifies oxygen consumption, digestive process and growth of an intertidal fish. Mar Environ Res 129:408–412
-
Gillett RM, White BN, Rolland RM (2008) Quantification and genetic profiling of DNA isolated from free-floating feces of the North Atlantic right whale (Eubalaena glacialis). Mar Mamm Sci 24:341–355
-
Goldberg CS, Pilliod DS, Arkle RS, Waits LP (2011) Molecular detection of vertebrates in stream water: a demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE 6:e22746
-
Goldberg CS, Sepulveda A, Ray A, Baumgardt J, Waits LP (2013) Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrus antipodarum). Freshw Sci 32:792–800
-
Goldberg CS, Turner CR, Deiner K, Klymus KE, Thomsen PF, Murphy MA et al (2016) Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol Evol 7:1299–1307
-
Gomez M, Arribas S, Vargas-Chacoff L (2016) Stress response of Salmo salar (Linnaeus 1758) facing low abundance infestation of Caligus rogercresseyi (Boxshall & Bravo 2000), an object in the tank, and handling. J Fish Dis 39:853–865
-
Günther B, Knebelsberger T, Neumann H, Laakmann S, Martínez Arbizu P (2018) Metabarcoding of marine environmental DNA based on mitochondrial and nuclear genes. Scientific Reports 8: 14822
-
Gustavson MS, Collins PC, Finarelli JA, Egan D, Conchúir RÓ, Wightman GD et al (2015) An eDNA assay for Irish Petromyzon marinus and Salmo trutta and field validation in running water. J Fish Biol 87:1254–1262
-
Haas C, Klesser B, Maake C, Bära W, Kratzera A (2009) mRNA profiling for body fluid identification byreverse transcription endpoint PCR and realtime PCR. Forensic Sci Int: Gen 3(2):80–88
-
Hall EM, Crespi EJ, Goldberg CS, Brunner JL (2016) Evaluating environmental DNA-based quantification of ranavirus infection in wood frog populations. Mol Ecol Resour 16:423–433
-
Hänfling B, Lawson Handley L, Read DS, Hahn C, Li J, Nichols P et al (2016) Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol Ecol 25:3101–3119
-
Hanson EK, Ballantyne J (2014) Rapid and inexpensive body fluid identification by RNA profiling-based multiplex High Resolution Melt (HRM) analysis. F1000Research, Version 2, 2:281
-
Harper LR, Handley LL, Hahn C, Boonham N, Rees HC, Gough KC et al (2018) Needle in a haystack? A comparison of eDNA metabarcoding and targeted qPCR for detection of the great crested newt (Triturus cristatus). Ecol Evol 8(12):6330–6341. https://doi.org/10.1002/ece3.4013
-
Havas M, Rosseland BO (1995) Response of zooplankton, benthos, and fish to acidification: an overview. Water Air Soil Pollution. 85:51–62
-
Hering D, Borja A, Jones JI, Pont P, Boets P, Bouchez A et al (2018) Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Res 138:192–205
-
Hunter ME, Oyler-McCance SJ, Dorazio RM, Fike JA, Smith BJ, Hunter CT et al (2015) Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive Burmese pythons. PLoS ONE 10:e0121655
-
Huver JR, Koprivnikar J, Johnson PTJ, Whyard S (2015) Development and application of an eDNA method to detect and quantify a pathogenic parasite in aquatic ecosystems. Ecol Appl 25:991–1002
-
Iversten LL, Kielgast J (2015) Monitoring of animal abundance by environmental DNA: an increasingly obscure perspective: a reply to Klymus et al., 2015. Biol Cons 192:479–480
-
Jane SF, Wilcox T, McKelvey K, Young MK, Schwartz MK, Lowe WH et al (2015) Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol Ecol Resour 15:216–227
-
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) "Sight-unseen" detection of rare aquatic species using environmental DNA. Conserv Lett 4:150–157
-
Jordahl DM, Benson A (1987) Effect of low pH on survival of brook trout embryos and yolk-sac larvae in West Virginia streams. Trans Am Fish Soc 116:807–816
-
Kelly RP (2016) Making environmental DNA count. Mol Ecol Resour 16:10–12
-
Kelly RP, Port JA, Yamahara KM, Crowder LB (2014) Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 9:e86175
-
Klymus K, Richter C, Chapman D, Paukert C (2015) Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol Cons 183:77–84
-
Knudsen SW, Bach Ebert RB, Hesselsøe M, Kuntke F, Hassingboe J, Bondgaard Mortensen P et al (2019) Species-specific detection and quantification of environmental DNA from marine fishes in the Baltic Sea. J Exp Mar Biol Ecol 510:31–45
-
Lacoursiere-Roussel A, Rosabal M, Bernatchez L (2016) Estimating fish abundance and biomass from eDNA concentrations: variability among capture methods and environmental conditions. Mol Ecol Resour 16:1401–1414
-
Laramie MB, Pilliod DS, Goldberg CS (2015) Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biol Cons 183:29–37
-
Levy D (1990) Sensory mechanism and selective advantage for diel vertical migration in juvenile sockeye salmon, Oncorhynchus nerka. Can J Fish Aquat Sci 47:1796–1802
-
Lintermans M, Raadik T, Morgan D, Jackcon P (2007) Overview of the ecology and impact of three alien fish speceis: redfin perch, Mozambique mouthbrooder (Tilapia) and Oriental weatherloach. In: Ansell D, and Jackson P, (eds) Emerging Issues in Alien Fish Management in the Murray-Darling Basin: Statement, recommendations and supporting papers. Proceedings of a workshop held in Brisbane QlD, May 2006 (pp 30–31) Murray-Darling Basin Commission, Canberra, AUS
-
Ma H, Stewart K, Lougheed S, Zhao J (2016) Characterization, optimization, and validation of environmental DNA (eDNA) markers to detect an endangered aquatic mammal. Conserv Genet Resour 8:561–568
-
Mächler E, Deiner K, Steinmann P, Altermatt F (2014) Utility of environmental DNA for monitoring rare and indicator macroinvertebrate species. Freshw Sci 33:1174–1183
-
Margurran AE (2004) Measuring biological diversity. Blackwell, Malden
-
Maron PA, Ranjard L, Mougel C, Lemanceau P (2007) Metaproteomics: a new approach for studying functional microbial ecology. Microb Ecol 53:486–493
-
Martellini A, Payment P, Villemur R (2005) Use of eukaryotic mitochondrial DNA to differentiate human, bovine, porcine and ovine sources in fecally contaminated surface water. Water Res 39:541–548
-
Maruyama A, Nakamura K, Yamanaka H, Kondoh M, Minamoto T (2014) The release rate of environmental DNA from juvenile and adult fish. PLoS ONE 5:e114639
-
Matsuhashi S, Doi H, Fujiwara A, Watanabe S, Minamoto T (2016) Evaluation of the environmental DNA method for estimating distribution and biomass of submerged aquatic plants. PLoS ONE 11:e0156217
-
McCormick SD, Moyes CD, Ballantyne JS (1989) Influence of salinity on the energetics of gill and kidney of Atlantic salmon (Salmo salar). Fish Physiol Biochem 6:243–254
-
McKenzie D, Hoglund E, Dupont-Prinet A, Larsen BK, Skov PV, Pedersen PB, Jokumsen A (2012) Effects of stocking density and sustained aerobic exercise on growth, energetics and welfare of rainbow trout. Aquaculture 338:216–222
-
Merkes CM, McCalla SG, Jensen NR, Gaikowski MP, Amberg JJ (2014) Persistence of DNA in carcasses, slime and avian faeces may affect interpretation of environmental DNA data. PLoS ONE 9:e113346
-
Milidine K, Armstrong J, Metcalfe N (1995) Presence of shelter reduces maintenance metabolism of juvenile salmon. Funct Ecol 20:839–845
-
Minamoto T, Uchii K, Takahara T, Kitayoshi T, Tsuji S, Yamanaka H, Doi H (2017) Nuclear internal transcribed spacer-1 as a sensitive genetic marker for environmental DNA studies in common carp Cyprinus carpio. Mol Ecol Resour 17:324–333
-
Moyer GM, Diaz-Ferguson E, Hill JE, Shea C (2014) Assessing environmental DNA detection in controlled lentic systems. PLoS ONE 9:e103767
-
Nakanishi H, Ohmori T, Hara M, Yoneyama K, Takada A, Saito K (2017) Identification of canine saliva using mRNA-based assay. Int J Legal Med 131:39–43
-
Ospina-Álvarez A, Palomera I, Parada C (2012) Changes in egg buoyancy during development and its effects on the vertical distribution of anchovy eggs. Fish Res 117–118:86–95
-
Parsons KM, Dallas JF, Claridge DE, Durban JW, Balomb KC, Thompson PM, Noble LR (1999) Amplifying dolphin mitochondrial DNA from faecal plumes. Mol Ecol 8:1766–1768
-
Person-Le Ruyet J, Mahe K, Le Bayon N, Le Delliou H (2004) Effects of temperature on growth and metabolism in a Mediterranean population of European sea bass, Dicentrarchus labrax. Aquaculture 237:269–280
-
Petty JT, Hansbarger JL, Huntsman BM (2012) Brook trout movement in response to temperature, flow, and thermal refugia within a complex Appalachian riverscape. Trans Am Fish Soc 141:1060–1073
-
Piaggio AJ, Engeman RM, Hopken MW, Humphrey JS, Keacher KL, Bruce WE, Avery ML (2014) Detecting an elusive invasive species: a diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Mol Ecol Resour 14:374–380
-
Piggott MP (2016) Evaluating the effects of laboratory protocols on eDNA detection probability for an endangered freshwater fish. Ecol Evol 6:2739–2750
-
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2013) Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can J Fish Aquat Sci 70:1123–1130
-
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2014) Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol Ecol Resour 14:109–116
-
Pochon X, Zaiko A, Fletcher LM, Laroche O, Wood SA (2017) Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications. PLoS ONE 12(11):e0187636
-
Pörtner HO (2010) Oxygen- and capacity-limitation of thermal tolerance: a matric for integrating climate-related stressor effects in marine ecosystems. J Exp Biol 213:881–893
-
Poté J, Ackermann R, Wildi W (2009) Plant leaf mass loss and DNA release in freshwater sediments. Ecotoxicol Environ Saf 72:1378–1383
-
Roberts D, Powell M (2005) The viscosity and glycoprotein biochemistry of salmonid mucus varies with species, salinity and the presence of amoebic gill disease. J Comp Physiol B 175:1–11
-
Robison BH, Bailey T (1981) Sinking rates and dissolution of midwater fish fecal matter. Mar Biol 65:135–142
-
Robson HLA, Noble TN, Saunders RJ, Robson SKA, Burrows DW, Jerry DR (2016) Fine-tuning for the tropics: application of eDNA technology for invasive fish detection in tropical freshwater ecosystems. Mol Ecol Resour 16:922–932
-
Roman J, McCarthy JJ (2010) The whale pump: marine mammals enhance primary productivity in a coastal basin. PLoS ONE 5:e13255
-
Sassoubre LM, Yamahara KM, Gardener LD, Block BA, Boehm AB (2016) Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ Sci Technol 50:10456–10464
-
Schmelzle MC, Kinziger AP (2018) Using occupancy modelling to compare environmental DNA to traditional field methods to regional-scale monitoring of an endangered aquatic species. Mol Ecol Resour 17:895–908
-
Scriver M, Marinich A, Wilson C, Freeland J (2015) Development of species-specific environmental DNA (eDNA) markers for invasive aquatic plants. Aquat Bot 122:27–31
-
Seidl MD, Pirow R, Paul RJ (2005) Acclimation of the microcrustacean Daphnia magna to warm temperatures is dependent on haemoglobin expression. J Therm Biol 30:532–544
-
Selong JH, McMahon TE, Zale AV, Barrows FT (2001) Effect of temperature on growth and survival of bull trout, with application of an improved method for determining thermal tolerance in fishes. Trans Am Fish Soc 130:1026–1037
-
Seymour M, Durance I, Cosby BJ, Ransom-Jones E, Deiner K, Ormerod SJ et al (2018) Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms. Commun Biol 1:4. https://doi.org/10.1038/s42003-017-0005-3
-
Shogren AJ, Tank JL, Andruszkiewicz EA, Olds B, Jerde C, Bolster D (2016) Modelling the transport of environmental DNA through a porous substrate using continuous flow-through column experiments. J R Soc Interface 13:20160290
-
Sloman K, Motherwell G, O'Connor KI, Tayor AC (2000) The effect of social stress on the standard metabolic rate (SMR) of brown trout, Salmo trutta. Fish Physiol Biochem 23:49–53
-
Somero GN (2002) Thermal physiology and vertical zonation of intertidal animals: optima, limits, and costs. Integr Comp Biol 42:780–789
-
Spear SF, Groves JD, Williams LA, Waits LP (2015) Using environmental DNA methods to improve detectability in a hellbender (Cryptobranchus alleganiensis) monitoring program. Biol Cons 183:38–45
-
Stewart K, Ma H, Zheng J, Zhao J (2017) Using environmental DNA to assess population-wide spatiotemporal reserve use. Conserv Biol 31:1173–1182
-
Stoeckle MY, Soboleva L, Charlop-Powers Z (2017) Aquatic environmental DNA detects seasonal fish abundance and habitat preference in an urban estuary. PLoS ONE 12:e0175186
-
Storch D, Santelices P, Barria J, Cabeza K, Pörtner HO, Fernández M (2009) Temperature tolerance of Zoea I from two different populations of the kelp crab Taliepus dentatus. J Exp Biol 212:1371–1376
-
Strickler KM, Fremier AK, Goldberg CS (2015) Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol Cons 183:85–92
-
Sundby S, Kristiansen T (2015) The principles of buoyancy in marine fish eggs and their vertical distributions across the world Oceans. PLoS ONE 10:e0138821
-
Sutherland WJ, Bardsley S, Clout M, Depledge MH, Dicks LV, Fellman L et al (2013) A horizon scan of global conservation issues for 2013. Trends Ecol Evol 28:16–22
-
Takahara T, Yamanaka H, Suzuki AA, Honjo MN, Minamoto T, Yonekura R et al (2011) Stress response to daily temperature fluctuation in common carp Cyprinus carpio L. Hydrobiologia 675:65–73
-
Takahara T, Minamoto T, Yamanaka H, Doi H, Kawabata Z (2012) Estimation of fish biomass using environmental DNA. PLoS ONE 7:e35868
-
Thaler J, McArt S, Kaplan I (2012) Compensatory mechanisms for ameliorating the fundamental trade-off between predator avoidance and foraging. Proc Natl Acad Sci USA 109:12075–12080
-
Thomsen PF, Kielgast J, Iversen LL, Wiuf C, Rasmussen M, Gilbert MTP et al (2012a) Monitoring endangered freshwater biodiversity using environmental DNA. Mol Ecol 21:2565–2573
-
Thomsen PF, Kielgast J, Iversen LL, Møller PR, Rasmussen M, Willerslev E (2012b) Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 7(8):e41732
-
Tirsgaard B, Svendsen JC, Steffensen JFL (2015) Effect of temperature on specific dynamic action in the Atlantic cod Gadus morhua. Fish Physiol Biochem 41:41–50
-
Treguier A, Paillisson J-M, Dejean T, Valentini A, Schlaepfer MA, Roussel J-M (2014) Environmental DNA surveillance for invertebrate species: advantages and technical limitations to detect invasive crayfish Procambarus clarkii in freshwater ponds. J Appl Ecol 51:871–879
-
Turner CR, Barnes MA, Xu CCY, Jones SE, Jerde CL, Lodge DM (2014) Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol Evol 5:676–684
-
Turner CR, Uy KL, Everhart RC (2015) Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol Conserv 183:93–102
-
Van der Marel M, Caspari N, Neuhaus H, Meyer W, Enss D, Steinhagen D (2010) Changes in skin mucus of common carp, Cyprinus carpio L., after exposure to water with a high bacterial load. J Fish Dis 33:431–439
-
Van Dievel M, Janssens L, Stoks R (2016) Short- and long-term behavioural, physiological and stoichiometric responses to predation risk indicate chronic stress and compensatory mechanisms. Oecologia 181:347–357
-
Vennemann M, Koppelkamm A (2010) mRNA profiling in forensic genetics I: possibilities and limitations. Forensic Sci Int 15:71–75
-
Watts P, Thompson DJ, Daguet C, Kemp S (2005) Exuviae as a reliable source of DNA for population-genetic analysis of odonates. Odonatalogica 34:183–187
-
Wilcox TM, McKelvey KS, Young MK, Lowe WH, Schwartz MK (2015) Environmental DNA particle size distribution from brook trout (Salvelinus fontinalis). Conserv Genet Resour 7:639–641
-
Wilcox TM, McKelvey KS, Young MK, Sepulveda AJ, Shepard BB, Jane SF, Whiteley AR, Lowe WH, Schwartz MK (2016) Understanding environmental DNA detection probabilities: a case study using a stream-dwelling char Salvelinus fontinalis. Biol Conserv 194:209–216
-
Wotton RS, Malmqvist B (2001) Feces in Aquatic Ecosystems: Feeding animals transform organic matter intofecal pellets, which sink or are transported horizontally by currents; these fluxes relocate organic matter inaquatic ecosystems. BioScience 51(7):537–544
-
Wright RF (2008) The decreasing importance of acidification episodes with recovery from acidification: An analysis of the 30-year record from Birkenes, Norway. Hydrol Earth Syst Sci 12:353–362
-
Wu RSS, Zhou BS, Randall DJ, Woo NYS, Lam PKS (2003) Aquatic hypoxia is an endocrine disruptor and impairs fish reproduction. Environ Sci Technol 37:1137–1141
-
Zubakov D, Kokshoorn M, Kloosterman A, Kayser M (2009) New marker for old stains: stable mRNA markers for blood and saliva identification of up to 16-year-old stains. Int J Legal Med 122:71–74
Acknowledgements
I would like to thank Maxinne Piggott for numerous discussions regarding this review. I would also like to thank Jacques Deere, Michelle Dileo, and Cam Hudson, for their input and Heidi Mumford for her indelible artistic aid. Finally, I would also like to thank the editor and two anonymous reviewers for their invaluable insights, ultimately improving the final version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Angus Jackson.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Reprints and Permissions
About this article
Cite this article
Stewart, K.A. Understanding the effects of biotic and abiotic factors on sources of aquatic environmental DNA. Biodivers Conserv 28, 983–1001 (2019). https://doi.org/10.1007/s10531-019-01709-8
-
Received:
-
Revised:
-
Accepted:
-
Published:
-
Issue Date:
-
DOI : https://doi.org/10.1007/s10531-019-01709-8
Keywords
- Aquatic conservation
- Biodiversity monitoring
- DNA production
- eDNA
- Genetic abundance
- Source dynamics
Source: https://link.springer.com/article/10.1007/s10531-019-01709-8
0 Response to "How Do Abiotic Factors Effect Marine Feeding Guilds"
Post a Comment